There is a new buzzword in the ocean science/sensing community. The word is eDNA, an abbreviation for environmental DNA. This refers to DNA that can be extracted from environmental samples without first isolating any target organisms. In the maritime community such samples are taken from water. All living organisms leave traces of DNA in their environments which is an indicator of their presence over time. This DNA is released into the environment through the biological process of living animals or by the decomposition of dead organisms. eDNA is a complex mixture of traces that enables the detection of a species regardless of its life stage or gender. This is a powerful tool for understanding the complex biological web of an ocean or coastal ecosystem.
While eDNA is potent, it is also ephemeral. Once released in the environment, the constituent DNA can be degraded by environmental factors. Generally, cold and dry conditions result in slow eDNA degradation. For example, in permafrost, eDNA can endure for hundreds or thousands of years. But in aquatic environments, the DNA released by an organism can be detected for only a few days. Despite this, the ocean science community is eager to employ this relatively new tool. In late November 2018 approximately 100 ocean scientists and stakeholders interested in marine eDNA assembled at The Rockefeller University in New York City for a conference sponsored by the Monmouth University-Rockefeller University (MURU) Marine Science and Policy Initiative. The executive summary of this gathering made it clear: “eDNA works. Get going.”
But what does that mean for technologists? How does this scientific method translate into operational ocean observing? Two research labs, The Monterey Bay Aquarium Research Institute (MBARI) in California and the Sieben Lab at Dalhousie University in Nova Scotia. among others, have been working to address the question.
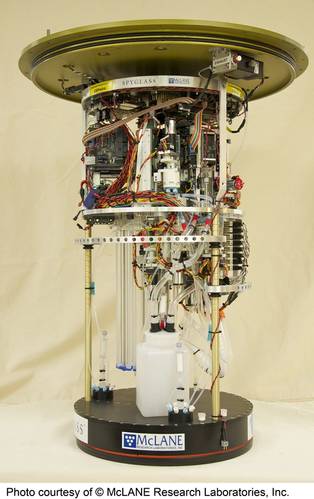
 A field-deployed microfluidic sensor (credit: Dartmouth Ocean Technologies Inc. and Sieben Laboratory Dalhousie University)MBARI has been working in this domain for decades. Starting about 25 years ago, it conceived of “ecogenomic sensors,” devices to apply molecular analytical techniques subsea. The Environmental Sample Processor (ESP) was conceived as an instrument to address the technological and operational concepts of ecogenomic sensors. Over time, the ESP emerged as a working instrument, enabling the use of DNA probes and protein arrays and the quantitative polymerase chain reaction (qPCR) technique to assess the presence and abundance of a range of organisms. The ESP is also used to preserve samples for a variety of laboratory tests not yet possible to carry out in situ. The instrument has been deployed on a variety of platforms, including coastal moorings, piers, an open ocean drifter, research vessels, a shallow water benthic lander, and a 4,000 m rated “elevator” designed for use on deep-sea cabled observatories.The initial motivation for developing the ESP was harmful algal blooms (HABs). Traditionally, understanding HABs required the transportation of physical field samples to a laboratory to identify the relevant organisms. This could take days or longer prompting the development of rapid methods for identifying and quantifying HABs of public health concern. Distinguishing toxic and nontoxic HAB species required species-specific molecular probes. But at the time it still took orders of magnitude longer to process samples using molecular probes as compared to physical, chemical, or optical measurements with commercially available sensors.
A field-deployed microfluidic sensor (credit: Dartmouth Ocean Technologies Inc. and Sieben Laboratory Dalhousie University)MBARI has been working in this domain for decades. Starting about 25 years ago, it conceived of “ecogenomic sensors,” devices to apply molecular analytical techniques subsea. The Environmental Sample Processor (ESP) was conceived as an instrument to address the technological and operational concepts of ecogenomic sensors. Over time, the ESP emerged as a working instrument, enabling the use of DNA probes and protein arrays and the quantitative polymerase chain reaction (qPCR) technique to assess the presence and abundance of a range of organisms. The ESP is also used to preserve samples for a variety of laboratory tests not yet possible to carry out in situ. The instrument has been deployed on a variety of platforms, including coastal moorings, piers, an open ocean drifter, research vessels, a shallow water benthic lander, and a 4,000 m rated “elevator” designed for use on deep-sea cabled observatories.The initial motivation for developing the ESP was harmful algal blooms (HABs). Traditionally, understanding HABs required the transportation of physical field samples to a laboratory to identify the relevant organisms. This could take days or longer prompting the development of rapid methods for identifying and quantifying HABs of public health concern. Distinguishing toxic and nontoxic HAB species required species-specific molecular probes. But at the time it still took orders of magnitude longer to process samples using molecular probes as compared to physical, chemical, or optical measurements with commercially available sensors.
Creating a portable “molecular biology laboratory in a can” was seen as a solution to more rapid HAB warnings following sample collection. After years of traditional lab techniques this was a non-trivial undertaking. By way of example, molecular analysis relies on use of perishable biochemical reagents, which are typically maintained in very precise conditions. Laboratory approaches can employ a clean, or even sterile, environment, have significant physical space, and are not power-limited. But the ESP overcomes these challenges. The ESP can perform qPCR unattended, while submerged. The instrument was commercialized and is now available from McLane Research Laboratories. This instrument, while functional, is larger and challenging to deploy.
More recently, a new version of the ESP has been developed for use aboard an autonomous underwater vehicle (AUV). In 2009 MBARI initiated an effort to re-engineer the ESP to fit on the long-range AUV, a novel AUV developed there. The new instrument, like its predecessor, was designed for both select sample analysis and sample preservation. Unlike its predecessors, however, the new ESP design concept is inherently more modular, with sample collection and handling distinct from sensors needed for real-time analytical work. This new ESP employs single-use cartridges containing both media for concentrating particles and reagents for processing that material. The interface between the cartridges and the sampler has a fixed design. Cartridges can evolve to meet new needs so long as they meet the interface specification. Two types of ESP cartridges have been devised. One is meant for sample preservation only, while the other is designed for homogenizing collected material and passage to downstream analytical modules. This cartridge based design has been tested at sea on the MBARI LRAUV.
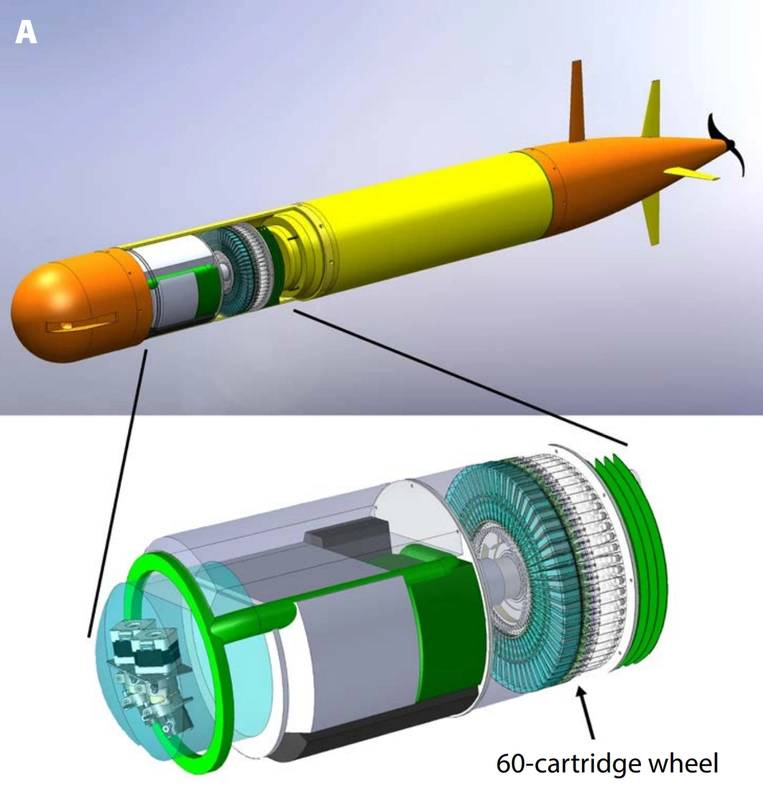 Illustration showing how the third-generation Environmental Sample Processor mounts inside MBARI’s long-range AUV (Credit: MBARI)
Illustration showing how the third-generation Environmental Sample Processor mounts inside MBARI’s long-range AUV (Credit: MBARI)
The use of microfluidics for in situ nutrient sensing has gained a great deal of traction in recent years. Microfluidic systems require less power, reagent, time, and human involvement than conventional instruments, making them ideal for long term deployments. These concepts are being employed to eDNA by the Sieben Lab at Dalhousie University in Nova Scotia. Their “Inlaid Optical Celltm” technology is a patent-pending approach that enables miniaturized optical spectroscopy on ultra-small amounts of fluid, less than a few microliters. Conserving reagent usage per measurement minimizes the volume of onboard chemicals and waste storage leading to smaller sensors.
These inlaid cells are used to perform absorbance and fluorescence measurements. In these systems, on-chip valving and pumping permit sophisticated fluid handling on the device, removing the need for expensive off-chip connectors and off-the-shelf solenoid valves and pumps. The on-chip valves also minimize dead or swept volume. The lab has developed an approach that uses magnetically tunable check valves for low-power and low-cost fluid routing solutions. By combining microfluidic valving, pumping, thermal control, and optical systems they can create lab-on-chip systems that automate long and complicated protocols to perform diverse chemistry and biomolecular assays. Examples of wet-chemistry methods include those for nutrients (nitrate, phosphate, silicate, etc.) and for genomics (integrated sample preparation, DNA capture/extraction, qPCR, etc.)
The team is collaborating with Dartmouth Ocean Technologies to make these sensors and instruments for deployment in remote environments. They have successfully demonstrated them on towed bodies and are commercializing new robust tools built on their microfluidic approach.
The field of eDNA monitoring is an emerging trend in ocean observing. As new instruments are refined and fielded the community will derive new insights about the behavior and distribution of marine life. This will be of utility to government agencies, such as fisheries regulators, and commercial operators such as fishers and aquaculture. There are also potential applications in defense and law enforcement. It is likely that by the end of this decade eDNA data will be as ubiquitous as the conductivity, temperature, and depth information that dominates physical oceanography today.






